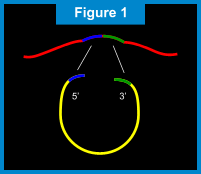
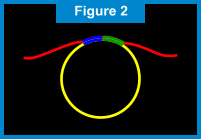
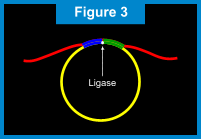
RAM - Ramification Amplication MethodIntroduction to RAMWith our RAM technology, a billion copies of a gene sequence can quickly and accurately be created (super-exponential amplification), in turn increasing discrimination power up to 1000 fold above current methods. Unlike other amplification techniques that require multiple cycles of alternating temperatures, RAM is an isothermal process that operates at a single temperature of 30°C. In actual clinical conditions, RAM is able to detect fewer than 5 targets and to produce one billion copies of that target in 1 hour*. This method is also used with extreme simplicity and sensitivity to detect viruses and bacteria in clinical specimens. In-situ (on a slide) detection of multiple viral sequences in cells is also possible using RAM technology. The ComponentsThe components required from RAM amplification include: 1) presence of a target nucleic acid sequence; 2) a circularizable probe; 3) a ligase; 4) DNA poylmerase; and 5) two extension primers, Primer 1 and Primer 2.  Return to TopHow RAM Works
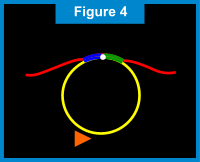
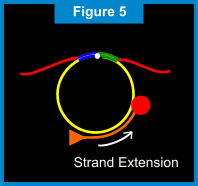
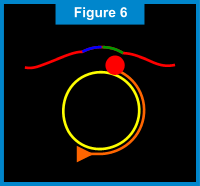
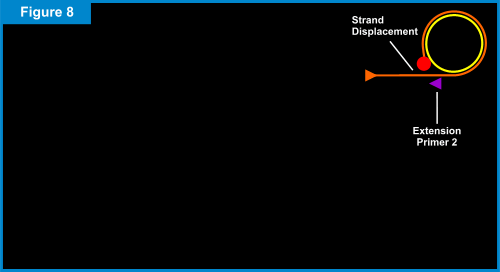
When the starting point is reached (e.g. one complete rotation around the circular probe), the newly formed Primer 1 strand is displaced incrementally and the DNA polymerase makes another trip around the circular probe. For each rotation around the circular probe, the displaced Primer 1 strand gets longer. (See Figure 8 below.) The newly displaced Primer 1 linear strand also acts as DNA a template. The second extension primer, Primer 2, which is complementary to a portion of this linear DNA template, binds to this linear DNA template. Once again, the DNA polymerase creates new nucleic acids along the template. As each rotation around the circular probe is completed, the linear DNA template gets longer allowing another Primer 2 to attach and extend. As the new Primer 2 linear strands extend, they eventually displace the "downstream" strands. This results in branching and creation of yet another set of linear DNA templates, to which the Primer 1 is complementary. Thus, Primer 1 attaches to the displaced Primer 2 linear strands and the process of extension, displacement and branching continues. (See Figures 9 & 10 below.) The end result of this amplification process is a super-exponential multiplication of the initial target sequence, which produces over a billion target sequences for easy detection.
Return to Top
|
||||||||||||||||