HSAM - Hybridization Signal Amplication MethodHSAM is an elegantly simple signal amplification method that takes advantage of some of the unique structural capabilities of nucleic acids. Conceptually, HSAM can be thought of as a variation on traditional probe network signal amplification schemes, updated by incorporation of cutting-edge nanostructure concepts. HSAM can be used to detect nucleic acids, proteins, or other small molecules that might typically be measured by immunoassay. In practice, a ligand-derivatized probe (nucleic acid or antibody) reacts specifically with the target molecule. Then, a self-assembling, structurally defined nanostructure, comprised of short ligand-derivatized nucleic acids and multivalent anti-ligand molecules, is introduced to bind to the target-bound probes. 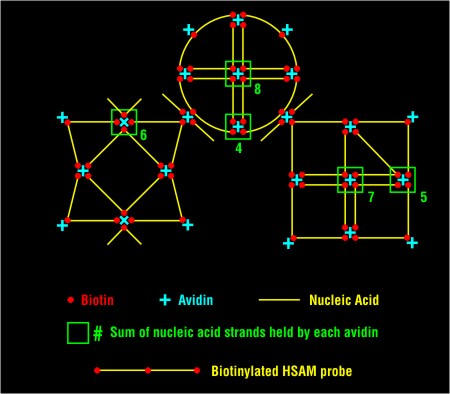 Components of the nanostructure can be derivatized with a variety of signal-generating moieties - e.g., fluorescent tags for direct detection or enzymes that can themselves generate detectable moieties in an indirect fashion - to suit specific applications. The nanostructure provides a many-fold greater surface area for supporting detection moieties than the probe molecule itself. HSAM is an attractive alternative to enzymatic amplification systems in situations where simplicity and speed are primary criteria in assay design. It should be particularly useful in solid phase or microarray types of applications where enzymatic amplification/detection can be cumbersome. HSAM also expands the signal amplification capabilities beyond nucleic acids to include proteins and other small antigens in the same analytical system. HSAM in conjunction with Hamilton Thorne BioScience's Ramification Amplification Method (RAM) constitutes a powerful family of amplification systems for a wide variety of applications. View a detailed technical narrative on HSAM (PDF)
|
||||
